Hi Guys!!
Welcome to my latest blog post, where I’ll be sharing the highlights of my journey in the Google Summer of Code (GSOC) program. Today, I’ll be covering the progress I made during the ninth week.
In my blog post for week 8, I outlined my plan to enhance the InMemoryDataset class for the breast cancer dataset. I also highlighted my intention to establish a validation split within the dataset and subsequently retrain my models, taking into account these new validation splits. During week 9, I progressed with these tasks.
My Progress in Week 9
Initially, I enhanced the InmemoryDataset class by streamlining it to incorporate solely the features and labels datasets. Unlike the ACC dataset, where separate classes were created for train and test splits, this is to enable users to determine their preferred split ratio when utilizing the dataset. Furthermore, to augment the class's utility, I integrated a custom split function within the class. This function enables users to effortlessly split the data, eliminating the need for external split functions such as Sklearn train_test_split. The class is displayed below:


To accomplish this, I revisited the preprocessing notebook for the brca_tcga dataset. Prior to any splitting, I saved the features and labels as separate CSV files. This process is illustrated below:

Finally, I uploaded all these to Zenodo to be able to get the download link that will be used in the InMemoryDataset class.
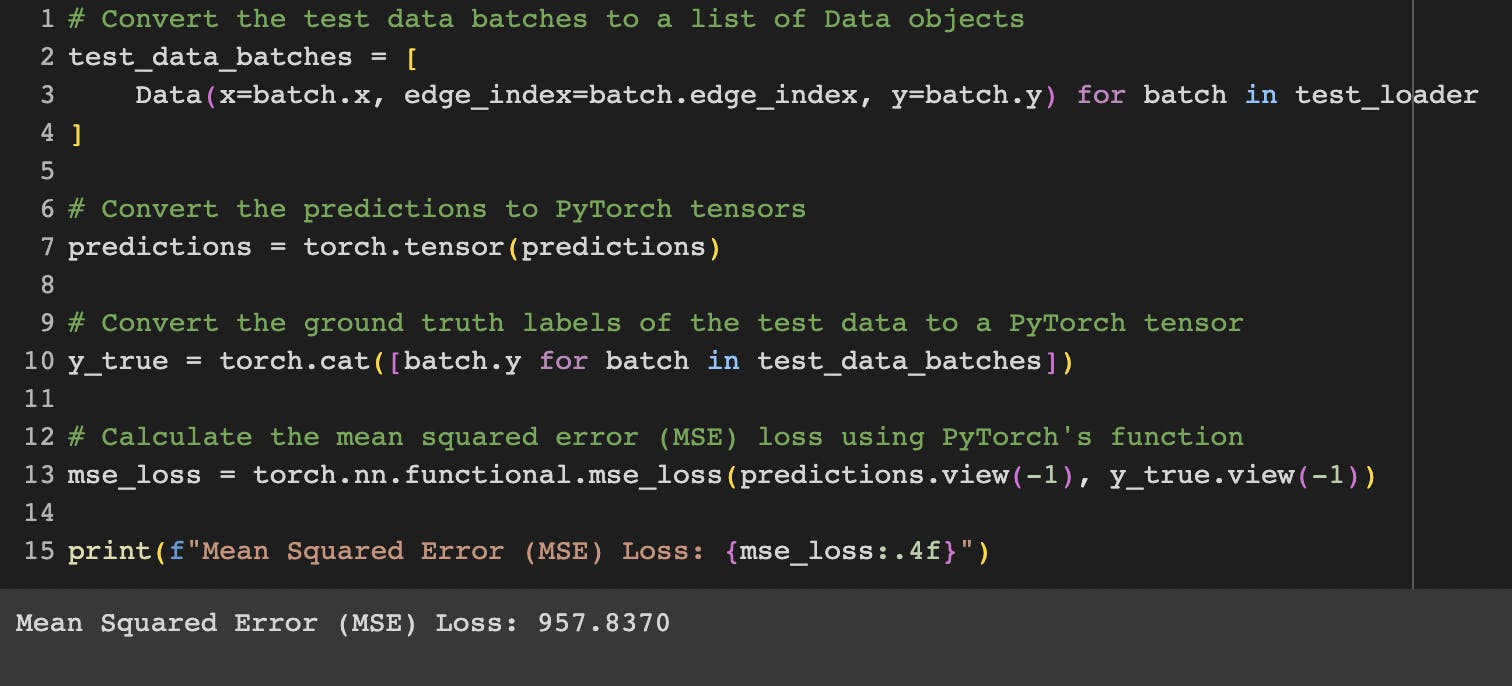
Moving on to the next task, I embarked on the process of training the Graph Neural Network (GNN) for the brca_tcga dataset. This involved utilizing the train and validation split for training purposes and subsequently conducting predictions on the test set. Encouragingly, the outcomes demonstrated superior performance in comparison to the baseline model generated through the utilization of FLAML. I also realized that for the ACC dataset, not utilizing GPU during model training led to lower performance scores compared to training with GPU acceleration. The Mean Squared Errors (MSEs) for these distinct models are respectively presented below:

Summary of Progress Made
Refined the InMemoryDataset class and included a custom split function for the brca_tcga dataset.
I worked on creating the GNN model for this dataset. I also worked on the baseline model for the dataset. Then, I was able to make comparisons between the two modelling techniques tried out. These results are been recorded in a spreadsheet that contains the dataset statistics which is seen below:

Challenges Faced
The primary challenge encountered this week was the constraint of available GPU resources on Colab during training. The limitation was evident in the inability to simultaneously train two notebooks utilizing GPU. Additionally, the substantial size of the breast cancer dataset would have depleted the daily GPU usage quota even after completing training for one notebook. This situation hindered the pace of my work progress. To address this challenge, I adopted a workaround involving the use of multiple email accounts to run notebooks concurrently. This approach required transferring both the notebook and associated data to the drive of the alternate email, allowing for more efficient use of GPU resources.
My Plans for Week 10
Develop a tailored Python script for PyG contribution, ensuring successful passage through all necessary tests.
Enhance the project notebooks by integrating mechanisms for acquiring essential data from Zenodo, bolstering the project's reproducibility for other users.
Prepare a research poster with my mentors to present at the upcoming Deep Learning Indaba 2023 conference, an event I am eagerly anticipating.
Overall, these past weeks have been incredibly productive, and the outcomes have been promising. The refinement of the InMemoryDataset class for the breast cancer dataset and the successful retraining of GNNs have marked significant steps forward in my project. Despite the challenges posed by GPU limitations, I'm pleased with the progress made.
I’m looking forward to sharing more updates and insights in the coming weeks. The upcoming tasks, including the development of a PyG-contributable Python script and enhancing notebook reproducibility, hold the potential to further elevate the project's impact. Your continued support and engagement have been invaluable throughout this exciting adventure!
I’m thrilled to share my progress with you all, and I invite you to join me for my next blog post. I’ll delve into the final stages of preparing the dataset for submission to PyTorch Geometric. Thank you for reading to the end!
(P.S. If you are interested in knowing more about my project, feel free to check it out on GitHub. https://github.com/cannin/gsoc_2023_pytorch_pathway_commons)