GSoC'23 Chronicles: Coding Week Six(6)
Creating torch_geometric.data.InMemoryDataset classes
Hi guys!!
Welcome to my latest blog post! Today, I'm excited to share my incredible progress during the sixth week of the Google Summer of Code (GSOC) program. I'm sharing these updates just now to catch up due to some unforeseen challenges that did not let me share the update for week 6 earlier.
In my previous blog post covering coding week 5, I mentioned that I would be focusing on making use of the InMemoryDataset class to wrap the dataset for contribution to PyTorch Geometric(PyG). This step was done to get familiar with this PyG class that is used when creating your own datasets. I also mentioned that I will be focusing on creating detailed notebooks to show the preprocessing for the ACC and breast cancer datasets gotten from the cBio portal and how they were integrated with the pathway commons dataset.
My Progress in Week 6
As mentioned in my post for weeks 3 and 4, I discussed briefly the InMemoryDataset class. This is an abstract class provided by PyG for setting up your own datasets ad it should be used if the whole dataset fits into memory.
In order to create a torch_geometric.data.InMemoryDataset, you need to implement four fundamental methods:
InMemoryDataset.raw_file_names(): A list of files inraw_dirwhich needs to be found in order to skip the download.InMemoryDataset.processed_file_names(): A list of files inprocessed_dirwhich needs to be found in order to skip the processing.InMemoryDataset.download(): Downloads raw data intoraw_dir.InMemoryDataset.process(): Processes raw data and saves it into theprocessed_dir.
Following the guidelines provided by PyG, I worked on creating the class for the ACC dataset. I created two different classes, one for the train set and one for the test set.
The raw files for the classes were the X_train, y_train, X_test, y_test and edge_index files which is show below:
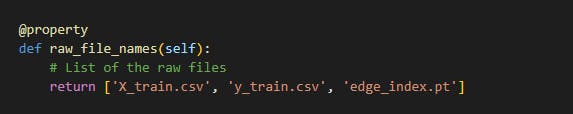
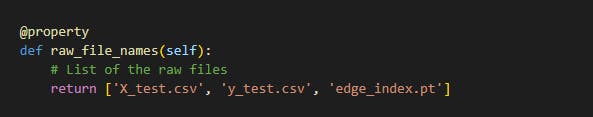
The processed files were the list of graphs generated from the train set and test set after processing it. The files are shown below:


For the download function, you have to specify a download link where the raw files will be downloaded. Before writing this function, your data has to be uploaded to a site where it can be downloaded from. I made use of the Zenodo site for this following the suggestion of my mentor. The zipped files of the train_set and test_set were uploaded to the site, then the download_url and extract_zip PyG functions were used in the function to download the files and unzip them.
def download(self):
# Download the file specified in self.url and store
# it in self.raw_dir
path = download_url(self.url, self.raw_dir)
extract_zip(path, self.raw_dir)
# The zip file is removed
os.unlink(path)
Finally, the process function is where the real work happens. In this function, a list of Data objects was created and saved into the processed_dir. Because saving a huge Python list is rather slow, we collate the list into one huge Data object via torch_geometric.data.InMemoryDataset.collate() before saving. The datasets were first loaded in, then processed to a list of patient-specific graphs which were finally converted to a list of data objects. The collated data object has concatenated all examples into one big data object and returns a slices dictionary to reconstruct single examples from this object. Finally, these two objects are loaded in the constructor into the properties self.data and self.slices.

During the week, I was also able to work on making the notebooks that show the preprocessing steps for the two datasets as detailed as possible. I did this by including different sections and more comments in the two different notebooks.

Summary of progress made
- Using
torch_geometric.data.InMemoryDataset: This step was done to set up the ACC dataset for more compatibility with PyG. It is a class with 4 different functions ad two different classes were created for the train set ad test set. The final output gotten from the classes are the list of graph data objects.

- Creating detailed notebooks for the preprocessing and integration of the datasets gotten from the cBio portal and Pathway Commons.
Challenges encountered
During the week, I encountered a significant challenge in determining the ideal platform for hosting and downloading my datasets. Initially, I attempted to utilize Google Drive, but unfortunately, the downloaded files were not in the correct format, causing inconveniences. Subsequently, I explored GitHub as an alternative option but encountered similar issues with incorrect file formats for the datasets.
I had a productive weekly meeting with my mentors, where I shared my concerns. They provided invaluable guidance and suggested employing Zenodo, an excellent platform capable of hosting various resources, including datasets. Following their advice, I uploaded my datasets to Zenodo, and it proved to be a remarkable solution. I was able to obtain the datasets in the desired format without any further complications.
My Plans for next week
For coding week 7, I will be focusing on creating baseline models for both the ACC dataset and the breast cancer dataset. To do this, my mentor suggested making use of automl models such as autokeras and FLAML.
Furthermore, I will also work on creating the torch_geometric.data.InMemoryDataset class for the breast cancer dataset. I will be discussing this with my mentors before proceeding in other to explore other functions of the PyG.
In Conclusion,
I am thrilled to present the exciting developments from my journey in GSoC coding week 6. Although this update arrives slightly later than anticipated, I am eager to promptly share the progress made in week 7. In the upcoming blog post, I will delve into the discussions and meetings I had with someone from PyG and my mentors. Additionally, I will show the remarkable results attained from the baseline models. Thank you for your unwavering support and for joining me on this weekly journey through my GSOC journey.